
Welcome here.
Learn everything you need to know about the much-talked-about NOBLEKINMAT BIOINFORMATICS DIPLOMA COURSE
Batch 8 registration is on-going
Registration closes 25th of January, 2026
Classes resume 1st of February, 2026

Noblekinmat Bioinformatics Diploma Course offers a comprehensive journey into the world of bioinformatics, focusing on genomics, transcriptomics, proteomics, data science, AI and computational biology.
Designed for students, researchers, and professionals, this program combines theoretical knowledge with practical skills, enabling participants to analyze complex biological data using cutting-edge tools and techniques.
Over the course of the program, learners will gain proficiency in essential bioinformatics methodologies, preparing them for impactful careers in research, healthcare, agriculture, environmental sciences, and biotechnology.
Whether you’re new to the field or looking to advance your expertise, this course provides the foundation and skills needed to excel in the dynamic world of bioinformatics.

𝐖𝐡𝐨 𝐒𝐡𝐨𝐮𝐥𝐝 𝐄𝐧𝐫𝐨𝐥𝐥?
1. Undergraduates/Graduates/Researchers: Gain the computational biology skills needed for advanced research.
2. Job Seekers: Prepare for bioinformatics roles in research, healthcare, or biotech.
3. Healthcare Professionals: Understand genomics and data analysis in precision medicine.
4. Freelancers: Offer bioinformatics services to labs and organizations.
5. Academics and Educators: Upgrade your teaching and research capacity with bioinformatics.
Batch 8 registration is on-going
Registration closes 25th of January, 2026
Classes resume 1st of February, 2026

Only one simple criteria
Have a computer and have access to the internet and electricity.
150+ students trained in 1 year
A proven curriculum designed by industry experts
Featured by leading education and research initiatives
A pathway to lucrative research, freelancing, and industry roles
Batch 8 registration is on-going
Registration closes 25th of January, 2026

Module 1: Molecular Biology and Introduction to Bioinformatics
⦁ DNA, RNA structure, and functions
⦁ Gene regulation, replication, and recombinant DNA technology
⦁ Evolution of genomes and bioinformatics applications
Module 2: Biological Databases and Tools
⦁ Major biological databases (NCBI, UniProt, PDB)
⦁ Sequence retrieval and analysis (e.g., BLAST)
⦁ Primer design using PrimerBank
Module 3: Sequence Alignment and Phylogenetics
⦁ Pairwise and multiple sequence alignment (Needleman-Wunsch, ClustalW)
⦁ Phylogenetic tree construction and molecular evolution studies
Module 4: Genome Analysis and NGS
⦁ Genome sequencing technologies
⦁ Assembly, annotation, and comparative genomics
⦁ NGS data analysis pipelines and tools
Module 5: Programming and Machine Learning in Bioinformatics I
⦁ Python, R, and Linux basics for bioinformatics
⦁ Workflow automation and hands-on coding exercises
Module 6: Structural Bioinformatics
⦁ Protein structure modeling (Swiss-model, RaptorX)
⦁ Drug design using AutoDock
⦁ Protein-ligand interaction studies
Module 7: Transcriptomics and Gene Expression Analysis
⦁ RNA-seq and differential gene expression analysis
⦁ Functional enrichment and pathway analysis
⦁ Gene regulatory networks
Module 8: Proteomics and Protein Function Analysis
⦁ Protein identification and annotation
⦁ Protein-protein interaction networks
⦁ Functional prediction tools
Module 9: Microbiome Research
⦁ Applications in medicine, agriculture, and environmental science
⦁ Microbiome analysis workflows
Module 10: Bioinformatics in Drug Discovery and Precision Medicine
⦁ Drug target identification and virtual screening
⦁ Pharmacogenomics and personalized medicine approaches
Module 11: Programming and Machine Learning in Bioinformatics II
⦁ Galaxy platform for bioinformatics
⦁ Introduction to machine learning models in bioinformatics
Module 12: Research Ethics and Writing
⦁ Principles of research ethics
⦁ Scientific writing and journal publishing
⦁ Journal club discussions
Module 13: Seminar and Project Presentation
⦁ Seminar presentations on selected topics
⦁ Final project showcasing practical applications of learned skills
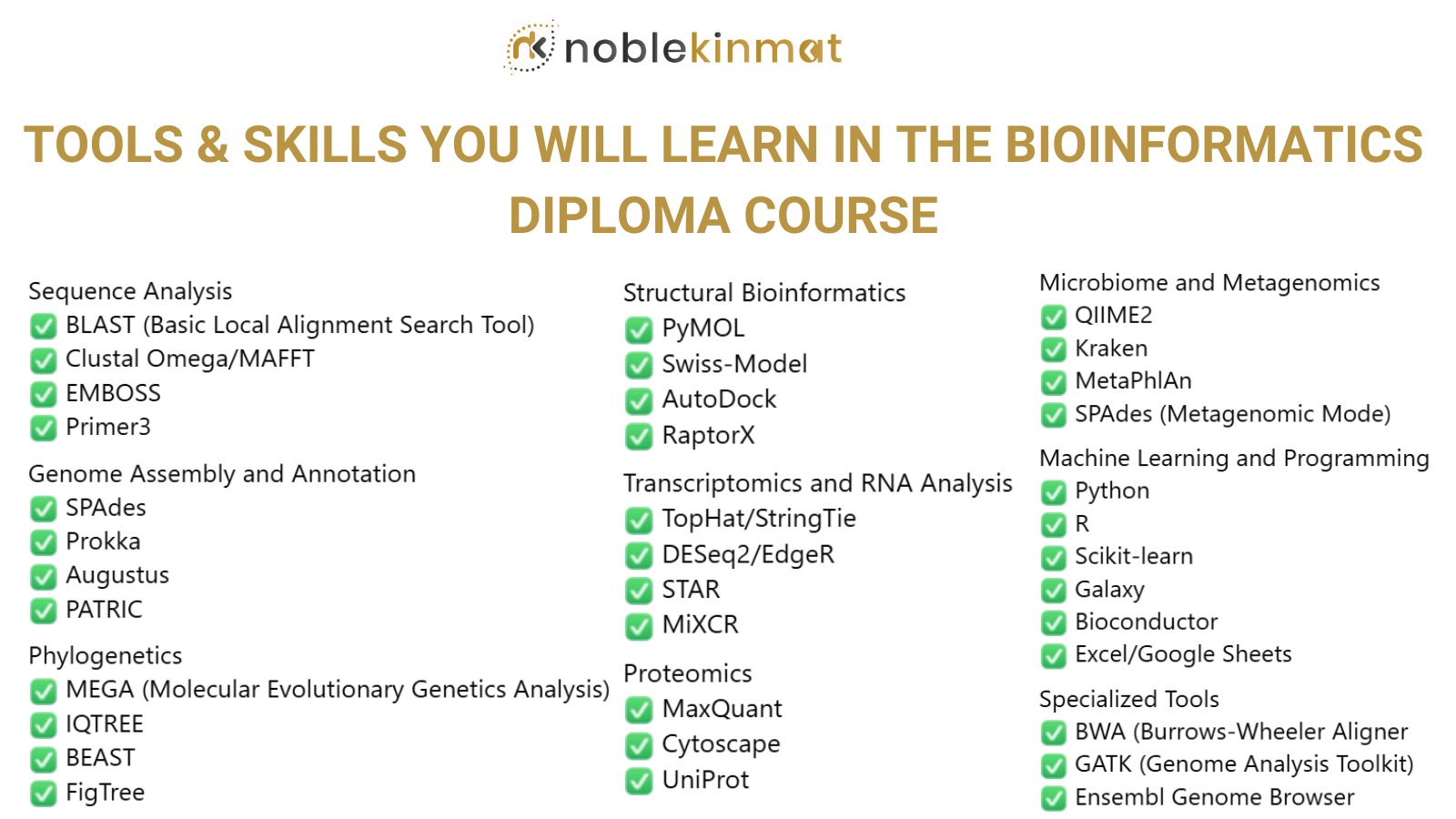
20+ Work-Ready Skills, Including:
✅Sequence Alignment and Analysis
✅Genome Annotation
✅NGS Data Analysis
✅Programming in Python/R
✅Phylogenetic Analysis
✅Data Visualization
✅Statistical Analysis
✅Database Management
✅Molecular Docking
✅Critical Thinking in Research
✅Scientific Report Writing
✅Presentation Skills
✅Project Management
✅Time Management
✅Freelancing in Computational Biology
✅Collaboration
✅Network Building
✅Ethical Research Practices
Batch 8 registration is on-going
Registration closes 25th of January, 2026
Classes resume 1st of February, 2026

Dr. Ashraf Akintola is a distinguished bioinformatics expert with a deep focus on genomic and microbiome research, particularly in mosquito vectors.
With a PhD in analyzing mitochondrial genomes and microbiome compositions, Dr. Akintola has made significant contributions to understanding evolutionary ecology and molecular adaptive evolution, especially in the context of climate change.
As the CEO of Noblekinmat Ltd (which includes Noblekinmat Bioinformatics), he leads initiatives in bioinformatics education, empowering students and professionals with cutting-edge skills in genomics, transcriptomics, and proteomics.
His expertise spans molecular phylogeny, endosymbiont studies, and the integration of AI in bioinformatics, making him a leading voice in advancing the field in Africa and beyond.
Check his research publications out here: https://scholar.google.com/citations?user=hCbzvD8AAAAJ&hl=en

𝐏𝐫𝐨𝐠𝐫𝐚𝐦 𝐃𝐞𝐭𝐚𝐢𝐥𝐬:
✅Flexible Learning: 100% online classes, accessible on mobile or laptop.
✅Beginner-Friendly: No prior bioinformatics experience is needed.
✅Live and Recorded Classes: 1-hour daily classes (Monday to Friday) and Occasional 2-hour live sessions on weekends from 8 pm to 10 pm WAT. All live sessions are recorded for convenience.
✅Interactive Support: Access to a vibrant learner community and mentorship from seasoned bioinformaticians.

All the tools you need to take your classes can easily be accessed on your phone to attend the classes but you need a laptop to do the analyses.
𝐏𝐫𝐨𝐠𝐫𝐚𝐦 𝐃𝐞𝐭𝐚𝐢𝐥𝐬:
1. Selar.co– A learning management software where your video classes are released daily.
2. Telegram – Just like Whatsapp, is used for the community. This is where everyone is free to share and connect. You would have access to materials and recordings there also.
You would not need to buy any software for all the analyses that would be taught.

Batch 8 registration is on-going
Registration closes 25th of January, 2026
Classes resume 1st of February, 2026

If you have any questions, please reach out to us at Noblekinmat@gmail.com
Course Objectives
By the end of the course, participants will:
⦁ Understand bioinformatics concepts and applications.
⦁ Analyze and interpret biological data using bioinformatics tools.
⦁ Perform genome and transcriptome analyses, including sequencing and annotation.
⦁ Develop skills in Linux, Python, and R programming for data handling.
⦁ Apply bioinformatics to solve real-world problems in genomics, precision medicine, and drug discovery.
Course Features
⦁ Duration: 8 weeks
⦁ Format: 100% online (live and self-paced learning)
⦁ Flexibility: Learn via pre-recorded videos and interactive live sessions.
⦁ Certification: Diploma awarded upon successful completion.
⦁ Prerequisites: Basic knowledge of biology; no prior programming experience required.
Learning Outcomes
Participants will be able to:
⦁ Perform sequence alignment, genome assembly, and annotation.
⦁ Conduct population genetics and phylogenetic analyses.
⦁ Analyze transcriptomics and proteomics data.
⦁ Use programming languages and machine learning techniques for bioinformatics challenges.
⦁ Apply bioinformatics skills to fields such as drug discovery, microbiome research, and precision medicine.
Copyright © NobleKinbio